My colleagues and I are estimating brain network activity in children using both neural and hemodynamic methods. These estimates will then be used to predict neurodevelopmental outcome later in life as quantified by Mullen and WPPSI tests. We expect that the combined brain network connectivity measures calculated using combined electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) will be more informative than metrics calculated from either modality alone. To accomplish this task, we are using an MRI-based localization scheme (inverse reconstruction) to project both modalities to a common brain space for each individual. The children in this project had EEG and fNIRS data collected with comparable baseline paradigms in two different sections.
We will compare the functional connectivity from both modalities within the same network(s) and between the same brain ROIs. The brain functional connectivity and network measures (e.g., graph theory) from both metrics will be associated with adverse experiences (e.g., SES and inflammation) and will be used to predict cognitive outcomes. The predictive power of each modality and class of metrics will be assessed separately and together.
Analysis plan
Datasets
The Bangladesh Early Adversity Neuroimaging (BEAN) dataset collected at 6, 24 and 36 months will be analyzed. This dataset was collected in a cohort of children exposed to early adversity as a consequence of growing up in an urban low-income neighborhood in Dhaka, Bangladesh. Task-free fNIRS and EEG were collected in each of these participants, and they also have neurodevelopmental outcomes measured at later time points (e.g., 24, 36 and 48 months). The BEAN dataset will be combined with the Neurodevelopmental MRI Database, which provides age appropriate MRIs, in order to do analyses for both EEG and fNIRS in brain space.
Connectivity profiles in EEG and fNIRS
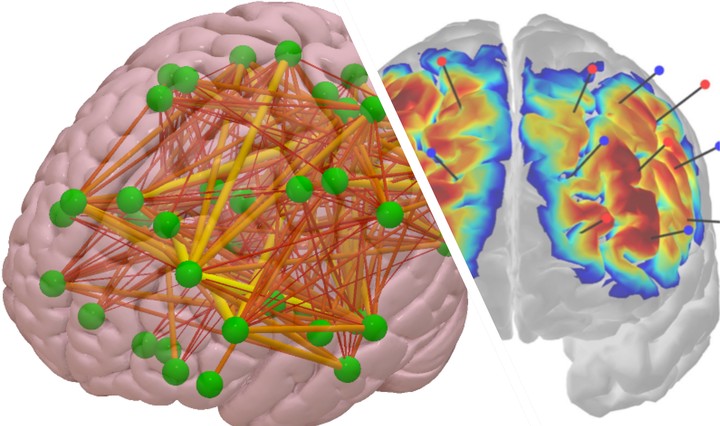
Brain functional connectivity analysis will be conducted with EEG and fNIRS data for each participant. The fNIRS functional connectivity will be estimated between the virtual channels on the scalp with corresponding 10-10 or 10-20 locations, as well as between cortical ROIs “localized” with the scalp-level fNIRS signals for both HbO and HbR signals using our recently developed inverse reconstruction methods (for fNIRS data). EEG functional connectivity will be estimated in different frequency bands (e.g., theta, alpha, beta, and gamma) between the 10-10, 10-20 locations corresponding to the fNIRS virtual channels on the scalp, as well as the cortical regions of interest (ROIs) in the brain that are reconstructed with EEG source analysis.
The inverse cortical reconstruction procedure will result in cortical activation in the same brain voxels (i.e., common space) for the two modalities. Inverse source reconstruction applies a spatial filter to signals recorded from the scalp to provide an estimate of the signal in “neural space.” Thus, the frame of analysis is no longer the external scalp location where the recording was done, but the cortical generators of scalp signal. One advantage of this method for the current project is that brain connectivity can be estimated from separately recorded EEG and fNIRS and reconstructed in a commensurable recording space, allowing for direct comparison in brain network activity between modalities.
Seed-based connectivity analysis will be conducted for both modalities using the same 10-10 locations and ROIs. The cortical areas detected by the fNIRS channels will be used as the “seeds”. The brain voxels/volumes in these seed areas will be used as the target sources for EEG connectivity analysis. As a result, cortical functional connectivity can be estimated for the two modalities (EEG and fNIRS) between the same brain ROIs for each participant.
Age-appropriate structural MRIs will be used for the localization of the fNIRS and EEG signals. An anatomical MRI close in head size and age will be selected for each Bangladeshi infant from the Neurodevelopmental MRI Database (Richards, Sanchez, Phillips-Meek, & Xie, 2016). Utilizing age-appropriate MRIs will improve the accuracy of the localization of EEG and fNIRS signals (Lloyd-Fox et al., 2014; Xie, Mallin, & Richards, 2018; Xie & Richards, 2017). This is especially important for infant participants, as changes in size, structure, and topology of the brain occur even within a narrowly defined age range.
Graph theory measures will finally be applied to capture the organization and overall architecture of the brain networks defined with EEG and fNIRS signals. Degree of nodes, betweeness centrality, and hub dependence will be measured to estimate the organization of the fNIRS and EEG brain networks, e.g., which brain areas are hubs in the brain functional networks during the task-free state. Path length, clustering coefficient, and small-worldness will be estimated to measure the local and global network efficiency in information transmission.